🚩 Tutorial¶
We’re going to look at some features of redflag, a library for helping find problems in machine learning pipelines.
You’ll need the following packages to run the code in this tutorial:
redflagpandasseaborn
A simple ML workflow¶
First, let’s see how we can burn ourselves:
X = [[19], [23], [35], [64], [59], [31]] # The smallest gamma-ray log.
y = ['ss', 'ss', 'ss', 'ms', 'ms', 'ss']
from sklearn.svm import SVC
clf = SVC(kernel='linear')
clf.fit(X, y)
clf.predict(X)
array(['ss', 'ss', 'ss', 'ms', 'ms', 'ss'], dtype='<U2')
So far so good. We’re predicting on the training data, but everything is at least working.
Now someone tells us we should scale our training data.
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler()
scaler.fit(X)
X_scaled = scaler.transform(X)
clf.fit(X_scaled, y)
clf.predict(X) # <-- Oops, we predicted on unscaled data.
array(['ms', 'ms', 'ms', 'ms', 'ms', 'ms'], dtype='<U2')
Easily done. There are lots of people on Stack Overflow and Cross Validated wondering why all their predictions are the same. It’s often because they’ve done something like this.
Even easier is this common pattern:
from sklearn.model_selection import train_test_split
scaler = StandardScaler()
scaler.fit(X)
X_train, X_test, y_train, y_test = train_test_split(X, y)
X_train_scaled = scaler.transform(X_train)
X_test_scaled = scaler.transform(X_test)
clf.fit(X_train_scaled, y_train)
clf.predict(X_test_scaled)
array(['ms', 'ss'], dtype='<U2')
There are at least three major problems with this block of code:
The split is totally random and not stratified to preserve the class imbalance in y.
The scaler was fit to the entire dataset, leaking test data into the model.
The data are correlated in a hidden feature (depth) and cannot be split randomly.
There are plenty of other problems too: it’s not reproducible, there’s not enough data, etc, etc.
These kinds of errors are everywhere in machine learning, and redflag wants to help change that.
A quick look at redflag¶
First make sure you have redflag v0.1.10 at least, otherwise do python -m pip install -U redflag in your environment.
import redflag as rf
rf.__version__
'0.5.0'
Load some data¶
import pandas as pd
df = pd.read_csv('https://raw.githubusercontent.com/scienxlab/datasets/main/kgs/panoma-training-data.csv')
df.head()
| Well Name | Depth | Formation | RelPos | Marine | GR | ILD | DeltaPHI | PHIND | PE | Facies | LATITUDE | LONGITUDE | ILD_log10 | Lithology | RHOB | Mineralogy | Siliciclastic | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | SHRIMPLIN | 851.3064 | A1 SH | 1.000 | 1 | 77.45 | 4.613176 | 9.9 | 11.915 | 4.6 | 3.0 | 37.978076 | -100.987305 | 0.664 | siltstone | 2393.499945 | siliciclastic | True |
| 1 | SHRIMPLIN | 851.4588 | A1 SH | 0.979 | 1 | 78.26 | 4.581419 | 14.2 | 12.565 | 4.1 | 3.0 | 37.978076 | -100.987305 | 0.661 | siltstone | 2416.119814 | siliciclastic | True |
| 2 | SHRIMPLIN | 851.6112 | A1 SH | 0.957 | 1 | 79.05 | 4.549881 | 14.8 | 13.050 | 3.6 | 3.0 | 37.978076 | -100.987305 | 0.658 | siltstone | 2404.576056 | siliciclastic | True |
| 3 | SHRIMPLIN | 851.7636 | A1 SH | 0.936 | 1 | 86.10 | 4.518559 | 13.9 | 13.115 | 3.5 | 3.0 | 37.978076 | -100.987305 | 0.655 | siltstone | 2393.249071 | siliciclastic | True |
| 4 | SHRIMPLIN | 851.9160 | A1 SH | 0.915 | 1 | 74.58 | 4.436086 | 13.5 | 13.300 | 3.4 | 3.0 | 37.978076 | -100.987305 | 0.647 | siltstone | 2382.602601 | siliciclastic | True |
For later use, I’m going to add a spurious column to the data:
import numpy as np
rng = np.random.default_rng(42)
df['Noise'] = rng.normal(size=len(df))
Imbalance metrics¶
redflag has some algorithms for various tasks, such as:
Imbalance metrics
Flagging data problems
Outlier detection
Distribution shape
Feature importance
Let’s look at imbalance first.
rf.imbalance_degree(df['Lithology'])
3.378593040846633
To interpret this number, split it into two parts:
The integer part, 3, is equal to \(m - 1\), where \(m\) is the number of minority classes.
The fractional part, 0.378…, is a measure of the amount of imbalance, where 0 means the dataset is balanced perfectly and 0.999… is really bad.
If the imbalance degree is -1 then there are no minority classes and all the classes have equal support.
In general, this statistic is more informative than the commonly used ‘imbalance ratio’ (rf.imbalance_ratio()), which is the ratio of support in the maximum majority class to that in the minimum minority class, with no regard for the support of the other classes.
We can get the minority classes, which are those with fewer samples than expected. These are returned in order, smallest first:
rf.minority_classes(df['Lithology'])
array(['dolomite', 'sandstone', 'mudstone', 'wackestone'], dtype='<U10')
Clipping¶
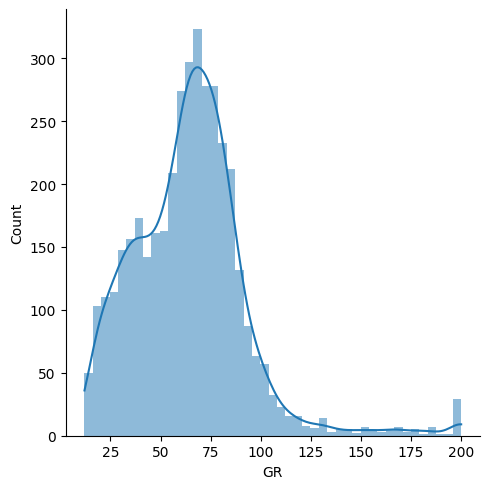
If a feature has been clipped, it will have multiple instances at its min and/or max value. There are legitimate reasons why this might happen, for example the feature may be naturally bounded (e.g. porosity is always greater than 0), or the feature may have been deliberately clipped as part of the data preparation process.
rf.is_clipped(df['GR'])
True
import seaborn as sns
sns.displot(df['GR'], lw=0, kde=True)
<seaborn.axisgrid.FacetGrid at 0x7fd7df18d760>
Independence assumption¶
If a feature is correlated to lagged (shifted) versions of itself (i.e. ‘autocorrelated’), then the dataset may be ordered by that feature, or the records may not be independent. For example, samples collected along a borehole are ordered by depth, and since the earth is organized, not random, this means that neighbouring samples will be correlated. Similarly, samples collected through time (the weather every hour) are often autocorrelated — you can predict the weather in an hour quite accurately by predicting it to be the same as the weather now.
If several features are correlated to themselves, then the data instances may not be independent, breaking the IID assumption.
Let’s see if our gamma-ray data are autocorrelated:
rf.is_correlated(df['GR'])
True
The property of being detectably autocorrelated is order-dependent. That is, shuffling the data apparently removes the correlation because the samples are now in random order, so any sample-to-sample correspondence appears to be gone:
import numpy as np
gr = df['GR'].to_numpy(copy=True)
np.random.shuffle(gr)
rf.is_correlated(gr)
False
But this does not mean the records are independent — only that you cannot tell that the records are autocorrelated.
A common way to deal with autocorrelation is to split the data differently. For example, if you have 1000 samples from 100 locations (or patients), with about 10 samples from each location (or patient), then it may be better (i.e. fairer) to split locations (or patients) into train and test sets, not samples. If you simply split samples, you will have records from inviduals locations (or patients) in both train and test, which is a common source of leakage.
Importance¶
We might like to see which of our features are more useful. There’s a function for that:
features = ['GR', 'RHOB', 'PE', 'Noise']
rf.feature_importances(df[features], df['Lithology'])
array([0.24637243, 0.19928635, 0.44311304, 0.11122817])
As we’d hope, the 'Noise' attribute is shown to be not very useful.
The relative importance of your features (dataset columns) for making accurate predictions is not a perfectly well-defined thing. Accordingly, there are several ways to measure feature importance. The feature_importances function aggregates three different measures of feature importance. The underlying models it uses depend on the type of task.
Classification tasks use the following:
A logistic regression model (using the absolute values of the coefficients).
A random forest classifier (based on mean reduction in impurity or Gini importance).
A K-nearest neighbours classifier (based on permutation feature importance with F1 score objective).
Regression tasks are assessed with the following:
A linear regression (using the absolute coefficients).
A random forest regressor, again using Gini importance.
A K-nearest neighbours, again with permutation importance but with mean squared error objective.
The aggregation function sums the normalized scores of the tests, and normalizes the result so that it sums to one.
Distributions¶
A common problem in the search for models is a mismatch between the distribution of the training and validation datasets. This might happen for several reasons, for example because of how the dataset is organized, how it was split, or because of how it was handled after splitting.
One simple error that can go unnoticed is fitting the model to scaled data, then forgetting to scale new data before prediction. Let’s see how redflag checks for this, with the wasserstein function.
wells = df['Well Name']
features = ['GR', 'RHOB', 'ILD_log10', 'PE']
w = rf.wasserstein(df[features], groups=wells, standardize=True)
w
array([[0.25985545, 0.28404634, 0.49139232, 0.33701782],
[0.22736457, 0.13473663, 0.33672956, 0.20969657],
[0.41216725, 0.34568777, 0.39729747, 0.48092099],
[0.0801856 , 0.10675027, 0.13740318, 0.10325295],
[0.19913347, 0.21828753, 0.26995735, 0.33063277],
[0.24612402, 0.23889923, 0.26699721, 0.2350674 ],
[0.20666445, 0.44112543, 0.16229232, 0.63527036],
[0.18187639, 0.34992043, 0.19400917, 0.74988182],
[0.31761526, 0.27206283, 0.30255291, 0.24779581]])
Pipelines¶
To make things as easy as possible, it would be nice to have some smoke alarms in the pipeline. Redflag has some prebuilt smoke alarms, and you can also make your own.
Redflag’s smoke alarms won’t be able to catch everything, however. For example if the data are shuffled and/or randomly sampled in a split, it might be very hard to spot self-correlation. I’m not sure how to alert the user to that kind of error, other than by potentially providing a wrapped version of train_test_split().
Anyway, let’s split our data in a sensible way: by well.
features = ['GR', 'RHOB', 'PE', 'Noise']
test_wells = ['CRAWFORD', 'STUART']
test_flag = df['Well Name'].isin(test_wells)
X_test = df.loc[test_flag, features]
y_test = df.loc[test_flag, 'Lithology']
X_train = df.loc[~test_flag, features]
y_train = df.loc[~test_flag, 'Lithology']
rf.pipeline
Pipeline(steps=[('rf.imbalance', ImbalanceDetector()),
('rf.clip', ClipDetector()),
('rf.correlation', CorrelationDetector()),
('rf.multimodality', MultimodalityDetector()),
('rf.outlier', OutlierDetector()),
('rf.distributions', DistributionComparator()),
('rf.importance', ImportanceDetector()),
('rf.dummy', DummyPredictor())])In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
Pipeline(steps=[('rf.imbalance', ImbalanceDetector()),
('rf.clip', ClipDetector()),
('rf.correlation', CorrelationDetector()),
('rf.multimodality', MultimodalityDetector()),
('rf.outlier', OutlierDetector()),
('rf.distributions', DistributionComparator()),
('rf.importance', ImportanceDetector()),
('rf.dummy', DummyPredictor())])ImbalanceDetector()
ClipDetector()
CorrelationDetector()
MultimodalityDetector()
OutlierDetector()
DistributionComparator()
ImportanceDetector()
DummyPredictor()
We can include this in other pipelines:
from sklearn.pipeline import make_pipeline
pipe = make_pipeline(StandardScaler(), rf.pipeline, SVC())
pipe
Pipeline(steps=[('standardscaler', StandardScaler()),
('pipeline',
Pipeline(steps=[('rf.imbalance', ImbalanceDetector()),
('rf.clip', ClipDetector()),
('rf.correlation', CorrelationDetector()),
('rf.multimodality', MultimodalityDetector()),
('rf.outlier', OutlierDetector()),
('rf.distributions', DistributionComparator()),
('rf.importance', ImportanceDetector()),
('rf.dummy', DummyPredictor())])),
('svc', SVC())])In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
Pipeline(steps=[('standardscaler', StandardScaler()),
('pipeline',
Pipeline(steps=[('rf.imbalance', ImbalanceDetector()),
('rf.clip', ClipDetector()),
('rf.correlation', CorrelationDetector()),
('rf.multimodality', MultimodalityDetector()),
('rf.outlier', OutlierDetector()),
('rf.distributions', DistributionComparator()),
('rf.importance', ImportanceDetector()),
('rf.dummy', DummyPredictor())])),
('svc', SVC())])StandardScaler()
Pipeline(steps=[('rf.imbalance', ImbalanceDetector()),
('rf.clip', ClipDetector()),
('rf.correlation', CorrelationDetector()),
('rf.multimodality', MultimodalityDetector()),
('rf.outlier', OutlierDetector()),
('rf.distributions', DistributionComparator()),
('rf.importance', ImportanceDetector()),
('rf.dummy', DummyPredictor())])ImbalanceDetector()
ClipDetector()
CorrelationDetector()
MultimodalityDetector()
OutlierDetector()
DistributionComparator()
ImportanceDetector()
DummyPredictor()
SVC()
pipe.fit(X_train, y_train)
🚩 The labels are imbalanced by more than the threshold (0.420 > 0.400). See self.minority_classes_ for the minority classes.
🚩 Features 0, 1 have samples that may be clipped.
🚩 Features 0, 1, 2 have samples that may be correlated.
🚩 Feature 0 has a multimodal distribution.
ℹ️ Multimodality detection may not have succeeded for all groups in all features.
🚩 There are more outliers than expected in the training data (316 vs 31).
🚩 Feature 3 has low importance; check for relevance.
ℹ️ Dummy classifier scores: {'f1': 0.2597634475275152, 'roc_auc': 0.4976020599836386} (stratified strategy).
Pipeline(steps=[('standardscaler', StandardScaler()),
('pipeline',
Pipeline(steps=[('rf.imbalance', ImbalanceDetector()),
('rf.clip', ClipDetector()),
('rf.correlation', CorrelationDetector()),
('rf.multimodality', MultimodalityDetector()),
('rf.outlier',
OutlierDetector(threshold=3.643721188696941)),
('rf.distributions', DistributionComparator()),
('rf.importance', ImportanceDetector()),
('rf.dummy', DummyPredictor())])),
('svc', SVC())])In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
Pipeline(steps=[('standardscaler', StandardScaler()),
('pipeline',
Pipeline(steps=[('rf.imbalance', ImbalanceDetector()),
('rf.clip', ClipDetector()),
('rf.correlation', CorrelationDetector()),
('rf.multimodality', MultimodalityDetector()),
('rf.outlier',
OutlierDetector(threshold=3.643721188696941)),
('rf.distributions', DistributionComparator()),
('rf.importance', ImportanceDetector()),
('rf.dummy', DummyPredictor())])),
('svc', SVC())])StandardScaler()
Pipeline(steps=[('rf.imbalance', ImbalanceDetector()),
('rf.clip', ClipDetector()),
('rf.correlation', CorrelationDetector()),
('rf.multimodality', MultimodalityDetector()),
('rf.outlier', OutlierDetector(threshold=3.643721188696941)),
('rf.distributions', DistributionComparator()),
('rf.importance', ImportanceDetector()),
('rf.dummy', DummyPredictor())])ImbalanceDetector()
ClipDetector()
CorrelationDetector()
MultimodalityDetector()
OutlierDetector(threshold=3.643721188696941)
DistributionComparator()
ImportanceDetector()
DummyPredictor()
SVC()
pipe.predict(X_test)
🚩 Feature 0 has samples that may be clipped.
🚩 Features 0, 1, 2 have samples that may be correlated.
🚩 There are more outliers than expected in the data (26 vs 8).
🚩 Feature 2 has a distribution that is different from training.
array(['siltstone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'siltstone', 'sandstone', 'wackestone',
'wackestone', 'wackestone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'limestone', 'limestone', 'limestone',
'wackestone', 'wackestone', 'wackestone', 'wackestone',
'siltstone', 'siltstone', 'siltstone', 'mudstone', 'mudstone',
'mudstone', 'mudstone', 'mudstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'mudstone', 'wackestone', 'limestone',
'limestone', 'limestone', 'limestone', 'limestone', 'limestone',
'limestone', 'limestone', 'limestone', 'limestone', 'limestone',
'limestone', 'limestone', 'limestone', 'limestone', 'limestone',
'limestone', 'limestone', 'limestone', 'limestone', 'limestone',
'limestone', 'wackestone', 'wackestone', 'wackestone',
'wackestone', 'wackestone', 'wackestone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'wackestone', 'wackestone', 'wackestone',
'wackestone', 'wackestone', 'wackestone', 'wackestone',
'wackestone', 'wackestone', 'wackestone', 'wackestone',
'wackestone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'wackestone', 'wackestone', 'wackestone',
'wackestone', 'limestone', 'limestone', 'limestone', 'limestone',
'limestone', 'limestone', 'limestone', 'limestone', 'limestone',
'wackestone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'wackestone', 'wackestone', 'wackestone', 'wackestone',
'wackestone', 'wackestone', 'wackestone', 'wackestone',
'wackestone', 'wackestone', 'mudstone', 'wackestone', 'wackestone',
'siltstone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'siltstone', 'limestone', 'limestone',
'limestone', 'limestone', 'limestone', 'limestone', 'limestone',
'limestone', 'limestone', 'limestone', 'limestone', 'limestone',
'limestone', 'limestone', 'limestone', 'limestone', 'limestone',
'limestone', 'limestone', 'limestone', 'limestone', 'limestone',
'limestone', 'wackestone', 'wackestone', 'wackestone', 'siltstone',
'siltstone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'limestone', 'limestone', 'limestone', 'limestone',
'limestone', 'limestone', 'limestone', 'limestone', 'limestone',
'wackestone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'limestone', 'limestone', 'limestone',
'limestone', 'limestone', 'limestone', 'limestone', 'limestone',
'limestone', 'wackestone', 'limestone', 'siltstone', 'siltstone',
'siltstone', 'limestone', 'limestone', 'limestone', 'limestone',
'limestone', 'limestone', 'limestone', 'limestone', 'limestone',
'siltstone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'mudstone', 'wackestone', 'wackestone',
'limestone', 'limestone', 'limestone', 'limestone', 'limestone',
'limestone', 'limestone', 'limestone', 'limestone', 'limestone',
'siltstone', 'limestone', 'mudstone', 'mudstone', 'wackestone',
'wackestone', 'wackestone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'siltstone', 'limestone', 'limestone',
'limestone', 'limestone', 'limestone', 'limestone', 'limestone',
'limestone', 'limestone', 'limestone', 'limestone', 'limestone',
'limestone', 'limestone', 'limestone', 'limestone', 'limestone',
'limestone', 'limestone', 'limestone', 'limestone', 'limestone',
'wackestone', 'wackestone', 'wackestone', 'wackestone',
'wackestone', 'wackestone', 'wackestone', 'wackestone',
'wackestone', 'wackestone', 'wackestone', 'wackestone',
'wackestone', 'wackestone', 'wackestone', 'wackestone',
'wackestone', 'wackestone', 'wackestone', 'wackestone',
'wackestone', 'wackestone', 'wackestone', 'wackestone',
'wackestone', 'wackestone', 'wackestone', 'wackestone',
'wackestone', 'wackestone', 'wackestone', 'wackestone',
'wackestone', 'wackestone', 'wackestone', 'wackestone',
'siltstone', 'siltstone', 'siltstone', 'mudstone', 'mudstone',
'mudstone', 'mudstone', 'mudstone', 'mudstone', 'mudstone',
'siltstone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'siltstone', 'wackestone', 'limestone',
'limestone', 'limestone', 'limestone', 'limestone', 'limestone',
'limestone', 'wackestone', 'wackestone', 'sandstone', 'sandstone',
'sandstone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'siltstone', 'wackestone', 'wackestone',
'siltstone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'wackestone', 'wackestone', 'limestone', 'limestone',
'limestone', 'limestone', 'limestone', 'wackestone', 'wackestone',
'wackestone', 'wackestone', 'wackestone', 'wackestone',
'wackestone', 'wackestone', 'wackestone', 'limestone',
'wackestone', 'wackestone', 'wackestone', 'wackestone',
'wackestone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'sandstone', 'sandstone', 'sandstone', 'sandstone',
'sandstone', 'sandstone', 'sandstone', 'sandstone', 'sandstone',
'sandstone', 'sandstone', 'sandstone', 'sandstone', 'sandstone',
'sandstone', 'siltstone', 'siltstone', 'wackestone', 'wackestone',
'wackestone', 'wackestone', 'wackestone', 'wackestone',
'wackestone', 'wackestone', 'wackestone', 'wackestone',
'siltstone', 'siltstone', 'siltstone', 'siltstone', 'wackestone',
'wackestone', 'wackestone', 'siltstone', 'siltstone', 'sandstone',
'siltstone', 'limestone', 'limestone', 'limestone', 'limestone',
'limestone', 'limestone', 'limestone', 'limestone', 'limestone',
'limestone', 'limestone', 'limestone', 'limestone', 'wackestone',
'wackestone', 'wackestone', 'wackestone', 'wackestone',
'wackestone', 'wackestone', 'wackestone', 'limestone',
'wackestone', 'limestone', 'wackestone', 'wackestone',
'wackestone', 'limestone', 'limestone', 'limestone', 'wackestone',
'wackestone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'siltstone', 'siltstone', 'sandstone',
'siltstone', 'siltstone', 'limestone', 'limestone', 'limestone',
'limestone', 'limestone', 'limestone', 'limestone', 'limestone',
'limestone', 'limestone', 'limestone', 'limestone', 'limestone',
'limestone', 'siltstone', 'wackestone', 'mudstone', 'sandstone',
'sandstone', 'sandstone', 'limestone', 'siltstone', 'siltstone',
'mudstone', 'mudstone', 'mudstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'siltstone', 'siltstone', 'limestone',
'limestone', 'limestone', 'limestone', 'limestone', 'limestone',
'limestone', 'limestone', 'limestone', 'limestone', 'limestone',
'limestone', 'wackestone', 'wackestone', 'wackestone', 'siltstone',
'siltstone', 'siltstone', 'sandstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'limestone', 'limestone', 'limestone', 'limestone',
'limestone', 'limestone', 'limestone', 'limestone', 'wackestone',
'wackestone', 'wackestone', 'wackestone', 'wackestone',
'wackestone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'wackestone', 'wackestone', 'wackestone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'siltstone', 'limestone', 'limestone',
'limestone', 'limestone', 'limestone', 'limestone', 'wackestone',
'wackestone', 'limestone', 'limestone', 'limestone', 'limestone',
'limestone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'limestone', 'limestone', 'wackestone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'mudstone', 'limestone', 'mudstone', 'mudstone', 'siltstone',
'siltstone', 'siltstone', 'mudstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'siltstone', 'siltstone', 'siltstone',
'siltstone', 'siltstone', 'siltstone', 'siltstone'], dtype=object)
Making your own tests¶
from redflag import Detector
def has_negative(x) -> bool:
"""Returns True, i.e. triggers, if any samples are negative."""
return any(x < 0)
negative_detector = Detector(has_negative, "are negative")
pipe = make_pipeline(negative_detector, SVC()) # NB, no standardization.
pipe.fit(X_train, y_train)
🚩 Feature 3 has samples that are negative.
Pipeline(steps=[('detector',
Detector(func=<function BaseRedflagDetector.__init__.<locals>.<lambda> at 0x7fd7dcae7420>,
message='are negative')),
('svc', SVC())])In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
Pipeline(steps=[('detector',
Detector(func=<function BaseRedflagDetector.__init__.<locals>.<lambda> at 0x7fd7dcae7420>,
message='are negative')),
('svc', SVC())])Detector(func=<function BaseRedflagDetector.__init__.<locals>.<lambda> at 0x7fd7dcae7420>,
message='are negative')SVC()
The noise feature we added has negative values; the others are all positive, which is what we expect for these data.
(Careful! All standardized features will have negative values.)